Data collection
This study was approved by the Institutional Review Board of Yonsei University Gangnam Severance Hospital and Yonsei University Dental Hospital (IRB No. 3-2019-0062 & No. 2-2019-0031) and all research was carried out in accordance with relevant guidelines and regulations. This study was a retrospective study, for which the requirement for informed consent was waived by the Institutional Review Board of Yonsei University Gangnam Severance Hospital and Yonsei University Dental Hospital due to its data source and methods. The radiographs were randomly selected by two dentists from the archive of the Department of Conservative Dentistry, Yonsei University containing bitewing radiographs taken from January 2017 to December 2018 for caries diagnosis and treatment. Radiographs including only permanent teeth were used for data, with no additional information about the patients (e.g. sex, age, or other clinical information). We tried to include a variety of cases that were as similar as possible to those encountered in real-world clinical situations. Radiographs with low image quality, excessive distortion, or severe overlapping of proximal surfaces due to the anatomical arrangement of particular teeth were excluded, because those features would interfere with a precise caries diagnosis. The bitewing radiographs were taken with the aid of a film-holding device (RINN SCP-ORA; DENTSPLY Rinn, York, PA, USA) using a dental X-ray machine (Kodak RVG 6200 Digital Radiography System with CS 2200; Carestream, Rochester, NY, USA).
The collected data were transferred to a tablet (Samsung Galaxy Note 10.1; Samsung Electronics Co., Suwon, South Korea) as Digital Imaging and Communications in Medicine files, and the two well-trained observers (postgraduate students of the Department of Conservative Dentistry, with a minimum clinical experience of 5 years) examined the dental images sequentially. The observers were allowed to adjust the density or contrast of radiographs as they wished with no time limitation. The observers drew lines for the segmentation of dental structures (caries, enamel, dentin, pulp, metal restorations, tooth-colored restorations, gutta percha) (Fig. 1) on the bitewing radiographs. All types of dental caries (e.g., proximal, occlusal, root and secondary caries) that can be observed on bitewing radiographs were tagged regardless of the severity. Discrepancies in caries tagging (e.g., regarding the presence or absence of the caries and the size of the caries) were initially resolved by consensus between the two observers, and if the disagreement persisted, it was resolved by another author.
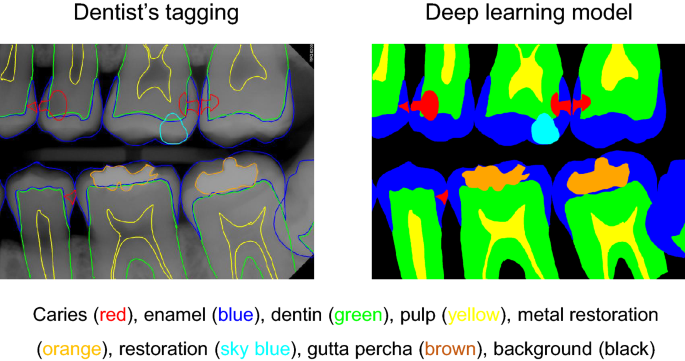
Example of the analysis of dental structures and caries tagging. The observers drew lines for the segmentation of dental structures (enamel, dentin, pulp, metal restoration, tooth-color restorations, gutta percha) and dental caries on the bitewing radiographs. *No software was used to generate the image. The picture used in Figure is a file printed using the source code that we implemented ourselves.
Training the convolutional neural network
The bitewing radiographs were directly used as diagnostic data for CNN without specific pre-processing (e.g. image enhancement and manual setting of the region of interest). The models were trained using each bitewing radiograph with 12-bit depth, which is the manufacturer’s raw format for bitewing radiographs, and its paired binary mask, while only the radiograph itself was fed into the models to detect the target regions for testing. The radiographs were scaled to the size of (572 times 572) to be used as input for the networks. In total, 304 bitewing radiographs, which were randomly divided into two groups (149 radiographs for ({mathcal{D}}_{{text{A}}}), which was used for training on both structure and caries segmentation, and 105 radiographs for ({mathcal{D}}_{B}), which was used for training on caries segmentation alone, as described below in greater depth) and a group of 50 radiographs with no dental caries (({mathcal{D}}_{{text{C}}} )), were used to train the deep learning model, while 50 radiographs (({mathcal{D}}_{{text{D}}} )) were used for performance evaluation. To evaluate the generality of the trained models without dataset selection biases, cross-validation was applied to the datasets ({mathcal{D}}_{{text{A}}} ,;{mathcal{D}}_{B}), and ({mathcal{D}}_{{text{C}}}). Each dataset was randomly split into six different folds exclusive of each other. The six folds were grouped into training, validation, and test sets at a 4:1:1 ratio for each repeat, with six rounds of repeated fold shuffling. In other words, each fold was not just a training set for a cross-validation model, but also a test set for the other cross-validation model. During the training time, the optimal model epoch was selected by evaluating the error rate of the radiographs of the validation set. After finishing the training, the test set, which was a totally independent fold from both the training and validation sets, was used to measure the final performance by using the model with the optimal epoch. If there had been pre-existing criteria for grading the level of difficulty of caries detection from the radiographs, we would have deliberately split the entire dataset into training, validation, and test sets according to those criteria. However, there are no standard criteria that could be used to gather each set without difficulty bias, which refers to the possibility that all the hard cases from the collected data could be included in the training set, whereas the easiest cases would be in the test set. Therefore, training and evaluating the model by randomly organized multiple cross-validation folds reduces the performance gaps between the test set and real-world data caused by dataset selection bias. Two augmentation processes were applied to the training set to train the model. Image augmentation, including intensity variation, random flipping, rotation, elastic transformation, width scaling, and zooming, was equally applied first to each radiograph and its paired binary mask. The image augmentation process acts as a regularizer that minimizes overfitting by randomly incorporating various image artifacts into the image dataset. Subsequently, mask augmentation consisting of random kernel dilations and elastic transformation was applied to only the binary masks. Mask augmentation was designed to reflect label inconsistencies among dentists. To detect the caries and structure regions from each input radiograph, this study used the U-Net architecture, a famous architecture for segmenting target regions on the pixel level. The U-Net architecture includes a convolutional part and an up-convolutional part. The convolutional part has the typical structure of convolutional neural networks with five convolutional layers. Contrastively, the up-convolutional part performs an up-sampling of the feature map by taking a concatenated output of the previous layer and the opposite convolutional layer as an input. Then, a (1 times 1) convolution maps the dimensionality of the feature maps to the desired number of classes. Finally, a softmax layer outputs a probability map where each pixel indicates the probability for each class within a range of (left[ {0, 1} right]) [ref. Softmax]. The network was optimized by the adaptive moment estimation optimizer with an initial learning rate of 0.00001 [ref. Adam optimizer]. Details on model architecture are provided in the Supplementary Fig. S1 online.
To accurately detect caries regions from the input bitewing radiographs, we designed a two-step process: detection and refinement. To do this, the U-Net was trained on two models: the U-Net for caries segmentation (U-CS), and the U-Net for structure segmentation (U-SS). The purpose of the U-CS is to extract carious segments from the input radiograph, whereas the U-SS segments dental structures. The input radiographs and their corresponding binary masks were used to train two U-Nets. The target regions for the U-CS and the U-SS were carious regions and structures, respectively. To train U-SS, the ({mathcal{D}}_{{text{A}}}) dataset was used and showed quite accurate performance in segmentation of dental structures with a small training dataset. In contrast, the U-CS was trained by consecutively accumulating the datasets from ({mathcal{D}}_{{text{A}}}) to ({mathcal{D}}_{C}) to observe the change in performance according to the features of the training dataset and the number of radiographs. Additionally, we introduced a penalty loss to overcome the limitation from the small scale of the training dataset, which can give rise to a large number of false detections. The penalty loss term was designed to assign a penalty to predicted carious regions on an input radiograph having no actual carious regions during training.
By simultaneously feeding each input radiograph into the U-SS and the U-CS, a caries probability map and a structure probability map were generated. The program was constructed to visualize detected dental caries as an area on the bitewing radiograph and also to show the degree of dental caries numerically. For the caries probability map, a pixel value of more than 0.55 was classified as a caries pixel. To find the threshold value of 0.55, we iteratively observed the agreement between the outputs of the trained model and experts’ diagnoses for the validation sets, while changing the threshold value from 0.01 to 0.99 with a step size of 0.01. In this process, the threshold value of 0.55 showed the optimal results in terms of agreement. To reduce the likelihood of false detection, areas of caries detected by the U-CS without overlap with enamel or dentin regions were eliminated. Figure 2 shows an overview of caries detection and false detection refinement.
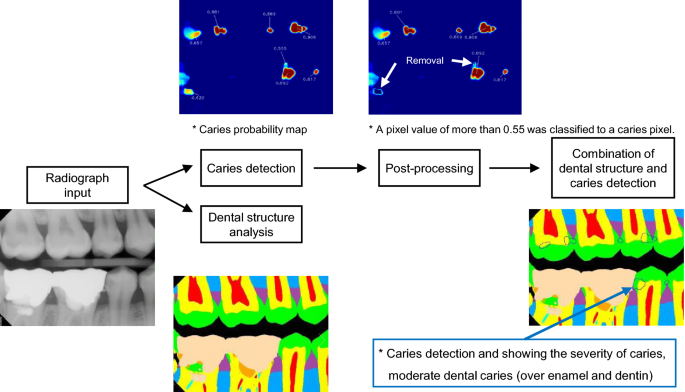
Flowchart of the detection of dental caries in the deep learning model, showing two models: the U-Net for caries segmentation (U-CS), and the U-Net for structure segmentation (U-SS). *No software was used to generate the image. The picture used in Figure is a file printed using the source code that we implemented ourselves.
Performance evaluation of the CNN model
To evaluate the performance of dental caries detection, the assessments were computed at the caries component level. If a blob classified as caries overlapped with a ground truth caries region at a specific ratio, we regarded the blob as a hit. We measured the precision (also known as positive predictive value (PPV), (TP/left( {TP + FP} right)), %), recall (also known as sensitivity, (TP/left( {TP + FN} right)), %), and F1-score (({text{F}}1;{text{score}} = {{left( {2left( {precision*recall} right)} right)} mathord{left/ {vphantom {{left( {2left( {precision*recall} right)} right)} {left( {precision + recall} right)}}} right. kern-nulldelimiterspace} {left( {precision + recall} right)}})) according to the overlap ratio ((theta)), which was set to 0.1, between the agreed-upon and predicted carious regions. We computed the final result by summing the number of caries from the test set of the cross-validation models on each dataset ({mathcal{D}}_{{text{A}}} ,{ }{mathcal{D}}_{{text{B}}} ,) and ({mathcal{D}}_{{text{C}}}), whereas ({mathcal{D}}_{{text{D}}}) was not included.
Performance evaluation and comparisons of dentists (before vs. after revision) with assistance of the CNN model
To assess whether the developed deep learning model can support the diagnosis of dentists, we used ({mathcal{D}}_{{text{D}}}), the evaluation dataset containing 50 radiographs. Specifically, we explored how dentists’ diagnoses changed before and after they referred to the predictions of dental caries made by the deep learning model. To do this, the deep learning model, which was previously trained with 304 training images, was used to detect dental caries in 50 radiographs, and three dentists (working in clinics with a clinical experience of 4–6 years) were instructed to tag dental caries independently, without any consultations. A few weeks later, the three dentists diagnosed the same radiographs, but with two distinguishable guidance lines, comprising the regions of dental caries predicted by the model and their previous diagnosis. The three dentists were instructed to revise (modify, delete, or add) their previously tagged dental caries by referencing the dental caries results detected by the deep learning model. As a result, each radiograph had seven sets of dental caries regions in total: six from dentists, and one from the model. To evaluate the validity of the model as a diagnostic support system, we analyzed the changes between first and second diagnoses of each dentist by measuring the PPV (%), sensitivity(%) and F1-score.Three dentists reached consensus on dental caries detection in ({ }{mathcal{D}}_{{text{D}}}), the evaluation dataset containing 50 radiographs, and divided the agreed-upon dental caries into 3 subgroups according to the severity of caries (initial, moderate, and extensive). The radiographic presentation of the proximal caries was classified as follows: sound: no radiolucency; initial: radiolucency may extend to the dentinoenamel junction or outer one-third of the dentin; moderate: radiolucency extends into the middle one-third of the dentin; and extensive: radiolucency extends into the inner one-third of the dentin1. If the same area was included, the same dental caries was considered to have been detected regardless of the overlap ratio on the caries lesion level; using this criterion, the sensitivity (%) was calculated according to the severity of the agreed-upon dental caries.
Statistical analysis
The diagnostic performance for readers and the U-Net CNN model was calculated in terms of the PPV (%), sensitivity (%), and F1-score. To compare the PPV and sensitivity between readers and the U-Net CNN model, generalized estimating equations (GEEs) were used, while the F1-score was compared using the bootstrapping method (resampling: 1000). All statistical analyses were performed using SAS (version 9.4, SAS Inc., Cary, NC, USA) and R package (version 4.1.0, http://www.R-project.org). The significance level was set at alpha = 0.05.

