We followed the “Strengthening the Reporting of Observational studies in Epidemiology” (STROBE) guidelines for observational studies66. All experiments were performed in accordance with the guidelines and regulations approved by the Institutional Review Board approval (HUM00105204 and HUM00113199) from the University of Michigan and the informed consent was obtained from all participants.
Study design, setting and participants
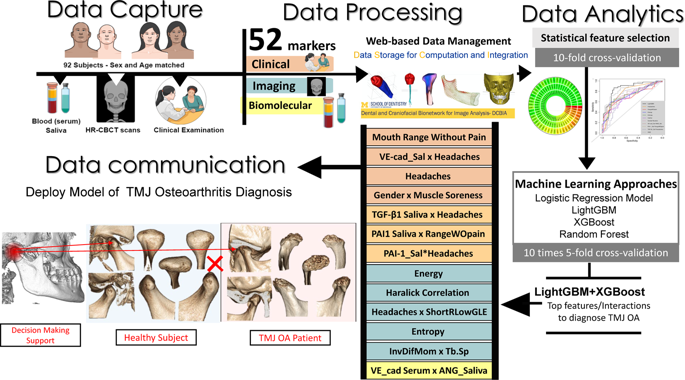
After the Institutional Review Board approval (HUM00105204 and HUM00113199) from the University of Michigan, we enrolled patients and subjects from January 2016 to December 2018 that composed our TMJ OA and Control groups, respectively. This cross-sectional study sample was composed of 92 patients, 46 TMJ OA and 46 age and sex-matched control subjects who were selected based on rigorous inclusion criteria. The general health conditions of the participants included: age between 21–70 years old, no history of cancer, no history of jaw joint trauma, no previous surgery in the TMJ or recent jaw joint injections, absence of systemic diseases; no current pregnancy and no congenital bone or cartilage disease. All patients were examined by a single temporomandibular disorders specialist at the Hospital of the University of Michigan (Medicine Oral Surgery Clinic) through the Diagnostic Criteria for Temporomandibular Disorders (DC/TMD)15 for TMJ osteoarthritis diagnosis. The patients were diagnosed as early stages of TMJ osteoarthritis when they presented: pain in at least one TMJ for less than 10 years, TMJ noise during movement or function in the last 30 days and crepitus detected during mandibular excursive movements. The Control group subjects were recruited by advertisement and evaluated for the absence of TMJ OA clinical and radiographic signs and symptoms. The diagnosis for the TMJ OA group and side of choice (left or right) was confirmed utilizing the radiographic criteria16, including initial stages of subchondral cyst, erosion, generalized sclerosis and/or osteophytes. For the matching control condyle, the side of choice was the one without any clinical or radiographic findings. The exclusion criteria for the TMJ OA group were patients with middle to chronic TMJ OA diagnosis, evaluated when they present more than 10 years of TMJ pain diagnosis and/or severe stages of bone destruction, subchondral cyst, erosion and generalized sclerosis evaluated using the hr-CBCT by a radiologist.
Variables
Our study was composed by 3 main sub-groups of variables, which were: biomolecular features (composed by proteins of serum and saliva), imaging features (composed by trabecular bone radiomics and morphometry) and clinical features.
Biomolecular data
We evaluated 14 proteins in serum and saliva associated with arthritis initiation and progression, such as nociception, inflammation, angiogenesis and bone resorption, which were: 6ckine, Angiogenin, BDNF, CXCL16, ENA-78, MMP-3, MMP-7, OPG, PAI-1, TGFb1, TIMP-1, TRANCE, VE-Cadherin and VEGF. However, the expression of 6ckine was not expressed in the serum and saliva samples in this study, and MMP-3 was not expressed in saliva. The raw data can be seen in the Supplementary Fig. 2. The reason to select those proteins, besides their participation in the TMJ OA inflammation process60, was due to our previous studies that detected these markers in the TMJ synovial fluid and saliva of OA patients, showing correlations with bone surface changes35,39.
Blood and saliva acquisition protocol
The participants had 5 ml of venous blood collected by a trained nurse at the University of Michigan. The blood was centrifuged for 20 minutes at 1000 RPM to separate only the serum that was then aliquoted in 2 ml Eppendorf tubes and stored at −80C. For the saliva collection, the participants received a 14 ml sterile test tube with a funnel inserted; they were instructed to tilt their head forward and drip the saliva off into the tube until 2 ml was collected. They were informed to not spit, talk, or swallow during this process67.
Custom micro-array
Custom human quantibody protein microarrays obtained from RayBiotech, Inc. Norcross, GA, was used to quantitatively assess the saliva and serum samples for the 14 specific biomarkers. Each participant had duplicates run for the saliva and serum samples (detailed description provided by Jiang et al.68 and Huang et al.69). Supplementary Figures 2, 3 shows the raw values obtained for each participant and the standard curves for each protein.
Clinical signs and symptoms acquisition protocol
The same investigator collected and measured the clinical signs and symptoms of the participants based on the DC/TMD15 criteria. The variables measured and selected for further statistical analysis were: Age pain began in years – TMJ OA Group only, Current Facial Pain -TMJ OA Group only, Worst Facial Pain in last 6 months -TMJ OA Group only, Average Pain -TMJ OA Group only, Last 6 Months Distressed by Headaches, Last 6 Months Distressed by Muscle Soreness, Vertical Range Unassisted Without Pain (mm), Vertical Range Unassisted Maximum (mm), Vertical Range Assisted Maximum (mm).
Imaging data acquisition
We acquired cone-beam computed tomography scans of each subject using the 3D Accuitomo (J. Morita MFG. CORP Tokyo, Japan) machine at the University of Michigan, School of Dentistry. The protocol for the temporomandibular joint high-resolution CBCT was field of view 40 × 40 mm; 90 kVp, 5 mAs, scanning time of 30.8 s and a voxel size of 0.08 mm3. The images were exported in DICOM (.dcm) using the manufacture software: i-Dixel (J. Morita MFG. CORP Tokyo, Japan) and optimization manufacture filter: G_103 + H_009. Finally, the images were coded and de-identified to avoid investigator bias in the statistical analysis.
Imaging trabecular texture-based features
We previously described the optimal parameters to extract radiomics features from the HR-CBCT scans in our study conditions and we followed these parameters to extract the information from our imaging data, using the BoneTexture module38. The region analyzed was the internal condylar lateral region (Fig. 6) due to our pilot results that showed this region to be the most significantly different between Control and TMJ OA patients. The textural information evaluated were: Energy, Entropy, Inverse Difference Moment, Inertia, Haralick Correlation, Short Run Emphasis, Long Run Emphasis, Grey Level Non Uniformity, Run Length Non Uniformity, Low Grey Level Run Emphasis, High Grey Level Run Emphasis, Short Run Low Grey Level Emphasis, Short Run High Grey Level Emphasis, Long Run Low Grey Level Emphasis, Long Run High Grey Level Emphasis, Bone Volume, Trabecular Thickness, Trabecular Separation, Trabecular Number and Bone Surface to Bone Volume Ratio.
Exploratory tests
We first did a traditional statistical analysis to explore our data and to test the hypothesis that there is no difference between our groups. Our data does not show normality distribution and for this reason, we chose non-parametrical tests for our analysis. The descriptive analysis, Mann-Whitney U test was done using the software GraphPad Prisma V 8.11 (GraphPad Software, Inc., San Diego, CA). For the descriptive analysis, we showed the median in addition to the mean, the 95% confidence intervals and the standard deviation. The Mann-Whitney U test was used to test our hypothesis and we used a two-tailed test with α of 5%.
Machine learning approaches
We diagnose the OA/control disease status based on the 52 features including five clinical variables, 20 radiomics features, 25 biomolecular features (13 from serum and 12 from saliva) and two demographic variables (age and gender). First, we normalized all features to have zero mean and one standard deviation. Next, we calculated the AUROC (Area under the Receiver Operating Characteristic curve), p-value and q-value70 from a two-sample Mann-Whitney U test to evaluate the significance of each feature (Fig. 3). Afterward, we compared four different prediction methods, each of which follows the four steps: (I) Cross-validation to avoid overfitting (II) feature selection (III) risk prediction (IV) method evaluation. We used one-sided paired DeLong test71,72 to validate the significance of AUC comparison between different approaches.
Cross-validation (CV)
We applied the 10 times’ 5-fold CV by taking 4 folds as training and the remaining one-fold as validation with 10 times’ repetition. At each time, we normalized the original 52 features denoted as F1 based on the training subjects and then took the product between each pair of them to generate additional 1326 interactions and denoted the set of 1378 features as ({{mathscr{F}}}_{2}). We performed the following two-step procedures by using only the training dataset and feature pools ({{mathscr{F}}}_{1}) and ({{mathscr{F}}}_{2}), respectively, where ({{mathscr{F}}}_{1}) represents the set of original 52 features, and we took the product between each pair of ({{mathscr{F}}}_{1}) to generate an additional 1326 interactions and denoted the set of 1378 features as ({{mathscr{F}}}_{2}). Afterwards, we applied the 10 times’ 5-fold CV by taking 4 folds as training and the remaining one-fold as validation with 10 times’ repetition. This will further evaluate the sensitivity of the model.
Feature selection
We calculate the AUC for each single feature in ({{mathscr{F}}}_{2}) and select top features according to {f ∈ F1|AUC of f > 0.7} and {f ∈ F2|AUC of f > 0.7} for feature pools met F1 and F2, respectively.
Evaluation and risk prediction
We trained the logistic regression model (method P1), Extreme Gradient Boosting (XGBoost; method ({{mathscr{P}}}_{2}))30, Light Gradient Boosting Machine (LightGBM; method ({{mathscr{P}}}_{3}))31, and Random Forest (method ({{mathscr{P}}}_{4}))73 model by using the extracted features from the last step for risk prediction of the validation subject. For both XGBoost and LightGBM models, we fix the depth D = 1, and tune the iteration steps by further splitting the training subjects into training and validation subjects for 10-fold cross validation, where AUC is chosen as the evaluation criterion. We evaluate the prediction performance of six pairs of feature set and methods (({{mathscr{F}}}_{1},,{{mathscr{P}}}_{1})), (({{mathscr{F}}}_{2},,{{mathscr{P}}}_{1})), (({{mathscr{F}}}_{2},,{{mathscr{P}}}_{2})), (({{mathscr{F}}}_{2},,{{mathscr{P}}}_{3})), (({{mathscr{F}}}_{2},,{{mathscr{P}}}_{4})) and (({{mathscr{F}}}_{2},,{{mathscr{P}}}_{2}+{{mathscr{P}}}_{3})) by using the accuracy, precision, recall, AUROC and ({{mathscr{F}}}_{1})-score74 on the 10 times 5-fold validation subjects. We also compare the results with other different hyperparameters. For example, we show in Table 4 the results for min_child_weight W∈{1,2}, colsample_bytree C∈{0.5,0.7}, subsample S∈ {0.5,0.7} and the learning rate η∈{0.001,0.01}. Our results showed that the XGBoost and LightGBM model by averaging the prediction probability (({{mathscr{F}}}_{2},,{{mathscr{P}}}_{2}+{{mathscr{P}}}_{3})) has the best performance on the validation subjects in the 10 times 5-fold CV; here the combination of XGBoost and LightGBM is recommended for its robustness in 10 times’ 5-fold CV.