Preparation and characterization of test agents
A NaF toothpaste (Colgate Triple Action, Colgate-Palmolive Company, USA), milled arginine (L-arginine monohydrochloride, Sigma-Aldrich, St. Louis, USA) and sterilized deionized water (DIW) were used to prepare fresh dentifrice slurry for biofilm treatment as per concentrations listed in Table 1. A standard ratio of 1:3 (dentifrice with/without arginine:DIW) was vortexed at 60 s at room temperature to prepare homogenized aqueous solution. The homogenized solution was then centrifuged at 4000 rpm for 20 mins (Hitachi high-speed centrifuge CR22N, Japan) at 25 °C. The sediment was discarded and the supernatant was transferred to a sterile tube for biofilm treatment and characterization34.
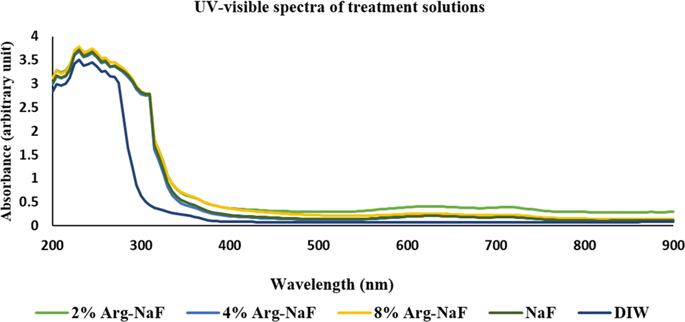
The freshly prepared test arginine-NaF solutions were analyzed for UV-visible characteristics because a notable change in color (as compared to NaF alone) was identified after incorporating the arginine. Randomly selected test solutions during different experimental phases were transferred to 96-well microplate to measure absorbance for UV-visible spectra (200–900 nm) at every 5 nm step increment using microplate reader35. The values thus obtained for each group-specific toothpaste supernatant was then summarized as mean to assess spectrum.
Bacterial strains and mono-species/3-species biofilm model
S. mutans UA159 (ATCC 700610), S. sanguis DSS-10 (ATCC 10556), and S. gordonii DL1 (ATCC 35105) were cultured at 37 °C under anaerobic condition (85% N2, 10% H2, 5% CO2) in brain heart infusion broth (BHI). Cell pellet of S. mutans, S. sanguinis, S. gordonii was adjusted to a concentration of 107 CFU/ml in BHI for biofilm growth.
Artificial saliva-coated hydroxyapaptite (sHA) discs (5 mm in diameter × 2 mm thickness; Clarkson Chromatography Products, Inc., South Williamsport, PA, USA) were used for biofilm growth. The disc dimensions were matched to estimate clinical dimensions of early enamel carious lesions. The manufacturer delivered HA discs were autoclaved and inserted in sterilized (autoclaved) artificial saliva (Phenol red, 4% NaOH, CaCl2, MgCl2.H2O, KH2PO4, KCl, HEPES, NaN3 – 1 ml/disc) for 1 h in incubator at 37 °C to simulate salivary pellicle formation. Mono-species and 3-species biofilm (inoculum ratio – 1:1:1 of S. mutans: S. sanguis: S. gordonii) were inoculated on sHA discs with BHI containing 1% sucrose (adjusted to pH 7) in anaerobic chamber (85% N2, 10% H2, 5% CO2) for 24 h. The biofilms were then dip-washed in sterile PBS (1 ml/disc) before receiving treatment with the test solutions. The treatment regimen for group-restricted test solutions (toothpaste supernatant) was: Group 1: 2% arginine – NaF (0.147% F) (2% Arg-NaF), Group 2: 4% arginine – NaF (0.144% F) (4% Arg-NaF), Group 3: 8% arginine – NaF (0.138% F) (8% Arg-NaF), Group 4 (toothpaste supernatant control): NaF (0.15% F) and Group 5: sterilized DIW. The biofilms were then transferred to the respective treatment solutions (180 µl/disc) for 1 min with constant agitation on a shaking incubator (80 rpm, 37 °C). The biofilms were then dip-washed in 1 ml/disc PBS for further transfer to wells containing BHI with 1% sucrose for 24 h anaerobic incubation (85% N2, 10% H2, 5% CO2). After 24 h, the biofilms were subjected to different characterizations. The group-specific mono-species biofilm were subjected to microbial cell viability assay, scanning electron microscopy (SEM) and confocal imaging. The 3-species biofilm were quantified for bacterial composition by DNA extraction and real-time polymerase chain reaction (PCR) analysis, SEM, confocal laser scanning microscopy, and RNA isolation for quantitative reverse-transcription real-time PCR analysis.
Microbial cell viability in mono-species biofilm
For each test group, inoculated treated biofilms on three sHA discs were transferred to a sterile tube with 1 ml PBS further vortexed for 60 s to receive biofilm suspensions. Aliquots of suspension were used for colorimetric microbial viability detection using a microbial viability assay kit (Dojindo Laboratories, Japan) – water-soluble tetrazolium salt (WST-8). Prior to assay, the coloring reagent was freshly prepared by mixing 9 parts of WST-8 solution with 1 part of diluted electron mediator reagent (mPMS) with dimethyl sulfoxide (DMSO). A 190-µl suspension aliquot was transferred to each well of a 96-well microplate followed by addition of 10-µl coloring reagent. The microplate was incubated for 2 h at 37 °C. After incubation, the absorbance was measured at 450 nm using microplate reader. The assay was performed in duplicate per biofilm suspension with repetitions at three time-points.
Scanning electron microscopy
Surface topographic biofilm (mono-species/3-species) assessment was done using SEM (SU1510, Hitachi, Japan) at 15 kV, 6000x. The biofilms were fixed with 2.5% glutaraldehyde in PBS (1 ml/disc) overnight. The biofilms were then serially dehydrated with graded series of ethanol concentrations (70%, 85%, 95%, 100%) at every ½ h. The biofilms were then air-dried at room temperature for 1 h, following which were subjected to sputtering (MSP-2S, IXRF systems, USA) with palladium and platinum (120 secs/disc). The sHA discs with inoculated biofilms were mounted on stubs for further analysis. Images subjected to qualitative assessment were captured at different randomly selected areas within the confines of the discs at 6000-x magnification.
Confocal imaging of biofilms
Confocal laser scanning microscopy (CLSM) of biofilms was done with CLS Biological Microscope (Olympus, FLUOVIEW FV1000, USA) – a two-photon laser scanning microscope. Prior to initializing scan, the treated biofilms were stained with LIVE/DEAD®BacLight™ Bacterial Viability Kit (SYTO 9/propidium iodide – 1:1 solution in DMSO) L7012 (Invitrogen detection technologies, USA) for 15 mins incubated at room temperature in dark. The biofilms on the sHA discs were transferred to circular coverslips mounted on the fluorescence microscope. Images were taken at 100-x magnification with three randomly selected view fields per biofilm. The images were then examined with Leica QWin v. 2.6. (Leica Microsystems Imaging Solutions, Germany) to determine the percent area with live and dead cells. A ratio of live/dead cells was calculated. The image examination to identify live/dead cells was done in triplicate per biofilm.
Total bacterial composition in 3-species biofilm
Biofilms were transferred to 1 ml PBS sterile tube to receive 60 s vortexed biofilm suspensions. The biofilm suspension was centrifuged at 7600 g for 10 mins. Then, the supernatant was decanted. The pellet was resuspended in 50 mM EDTA at pH 8.0 with 20 mg/ml lyzoyme and kept in water bath at 37 °C for 1 h. Then, the QIAamp Mini DNA kit (Qiagen, Hilden, Germany) was used to perform the DNA extraction as per manufacturer instructions. DNA extracted from S. mutans (ATCC 700610), S. sanguis (ATCC 10556) and S. gordonii (ATCC 35105) were used as the positive controls. The primers used for real-time PCR in the present study are listed in Table 2. Oligoneucleotide primers (Table 2) and probes (S. mutans P is 5′-FAM-TGGAAATGACGGTCGCCGTTATGAA-TAMRA-3′) were used (Applied Biosystems, USA). For each real-time PCR reaction (S. mutans), 20 µl of a mixture containing 5 µl of biofilm suspension, 1x Taqman Universal PCR Mix (Applied Biosystems, USA), 200 nM of forward (F) and reverse (R) primers and 250 nM Taqman Probe was prepared. The reaction was performed using Step One Plus (Applied Biosystems, USA). The cycle condition was: 50 °C/2 mins; 95 °C/10 mins; 50 cycles of 95 °C/15 sec and 58 °C/1 min. The standard curve was performed with the DNA of control ATCC S. mutans strain containing 5 × 103 – 1 × 107 CFU/ml. For each S. sanguis and S. gordonii real-time PCR reaction, 20 µl of a mixture containing 5 µl of biofilm suspension, 1x SYBR Green Master Mix (Applied Biosystems, USA) and 200 nM of F/R primers were used. The reaction was performed using Step One Plus (Applied Biosystems, USA). The cycle condition was: 95 °C/20 s; 50 cycles of 95 °C/3 s and 58 °C/30 s; and following melt-curve: 95 °C/15 s; 60 °C/1 min. The standard curve was performed with the DNA of control ATCC S. sanguis and S. gordonii strain containing 1 × 104 – 1 × 109 CFU/ml.
Relative target gene expression
Three sHA discs per test group inoculated biofilms were vortexed for 60 s in 1 ml of PBS to receive suspensions. The suspension was centrifuged at 7600 g for 10 mins. Then, the supernatant was decanted. The pellet was re-suspended in freshly-prepared TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.0) containing 0.4 mg/ml lyzoyme and kept in water bath at room temperature for 15 mins. Then, the SV Total RNA Isolation kit (Promega, Madison, USA) was used to extract the RNA as per manufacturer instructions. The purity of RNA was verified by absorbance measurement at A260/280 and run by gel electrophoresis. cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen detection technologies, USA) as per manufacturer instructions. Oligoneucleotide primers (Applied Biosystems, USA) as listed in Table 2 were used to quantify relative gene expression31. The relative quantification of gtfB, sagP, and arcA genes was performed using 16S rRNA as a reference gene; an internal control for quantification. For each real-time PCR reaction, 20 µl of a mixture containing 5 µl of suspension, 1x SYBR Green Master Mix (Applied Biosystems, USA) and 200 nM of F/R primers were used. The reaction was performed using Step One Plus (Applied Biosystems, USA). The cycle condition was: 95 °C/20 s; 50 cycles of 95 °C/3 s and 58 °C/30 s; and following melt-curve: 95 °C/15 s; 60 °C/1 min including appropriate negative and positive controls.
Statistical analysis
All experiments were repeated at least three times at disparate points. The statistical analysis of obtained data collocated in MS Office Excel 2016 (Microsoft Office Professional Plus 2016, Microsoft, USA) was done using SPSS v. 24 (IBM SPSS® Statistics Inc, USA). Data was assessed for normality and homogeneity of variances using Shapiro-Wilk test and Levene’s test, respectively. The statistical differences amongst the test groups for microbial cell viability was determined using Kruskal-Wallis One-way ANOVA with Dunn’s post-hoc test. The data for live/dead bacteria ratio was log transformed for homogeneity of variances and conversion to achieve the vital (live) cells existence subtracted by dead bacteria. The log-transformed data was further analyzed using one-way ANOVA followed by Student-Newman-Keuls (SNK) test for multiple comparisons. DNA and RNA quantified by real-time PCR in 3-species biofilm was subjected to parametric (factors: treatment and bacteria) and non-parametric (factors: treatment and gene) two-way ANOVA with post-hoc tests, respectively. The level of statistical significance for all tests was at 2-tailed p-value < 0.05.