In vitro cell culture experiments
In vitro cell culture experiments and methods were performed and reported as previously published.6,15,16,39,44 hPDL fibroblasts were obtained from the periodontal connective tissue of human teeth that were free of decay and were extracted for medical reasons. All experiments were performed in accordance with the relevant guidelines and regulations. Approval for the collection and usage of hPDL fibroblasts was obtained from the ethics committee of the University of Regensburg, Germany (approval number 12-170-0150). Briefly, we cultivated the tissue samples in six-well cell culture plates (37 °C, 5% CO2, 100% H2O) in complete media (high-glucose DMEM, D5796, Sigma-Aldrich®, St. Louis, MI, USA) with 10% FCS (P30-3306, PAN-Biotech, Aidenbach, Germany), 1% L-glutamine (SH30034.01, GE Healthcare Europe, Munich, Germany), 100 µmol·L−1 ascorbic acid (A8960, Sigma-Aldrich®) and 1% antibiotics/antimycotics (A5955, Sigma-Aldrich®) until proliferatory outgrowth of adherently growing fibroblasts was observed. The cells were characterised by hPDL-specific marker genes and a spindle-shaped morphology, as reported previously.15,44 For the in vitro study, hPDL fibroblasts from the third to fifth passages that were pooled from six individuals (three male, three female, ages 17–27 years) were seeded at a density of 2000 cells per mm2 into either standard six-well cell culture plates without oxygen permeability in the polystyrene base/membrane (353046, BD, Heidelberg, Germany) or in special gas-permeable Lumox® dishes (94.6077.331, Sarstedt, Nürnbrecht, Germany) with ultra-thin gas-permeable bases/membranes, which provided a continuous oxygen supply to the adherently growing fibroblasts at the base (Fig. 6).
Set-up used for the hPDL fibroblast experiments to evaluate the four experimental groups (1–4)
Experimental set-up
To simulate mechanical orthodontic strain in hPDL fibroblasts in compression areas of the periodontal ligament, a physiological orthodontic pressure of 2 g·cm–2 was applied under specific cell culture conditions (37 °C, 5% CO2, 100% water-saturated, 2 ml DMEM/well) for 48 h after a preincubation phase of 24 h by means of a sterilised glass disc according to an established and published method.6,7,15,16,39,44 (Fig. 6) The following four experimental groups with 6–9 biological replicates (samples) each (n) during 2–3 consecutive experiments (N) with three replicates each were incubated at 70% confluency for a total of 72 h: (1) no mechanical orthodontic compressive strain + normoxia (control, Lumox®); (2) mechanical strain + normoxia (Lumox®); (3) no mechanical strain + normoxia (control, polystyrene); (4) mechanical strain + hypoxia (polystyrene). In the conventional, non-gas-permeable polystyrene cell culture plates and in vivo, the applied compressive forces not only induced mechanical deformation and stress in the adherently growing hPDL fibroblasts but also limited the oxygen supply (group 4), which was not the case in the Lumox® plates with an intact oxygen supply via the gas-permeable membrane (group 2) (Fig. 6). This experimental set-up allowed for the experimental isolation and separation of the mechanotransducive and hypoxic effects that occur concomitantly during OTM, thus enabling an investigation of their respective importance.
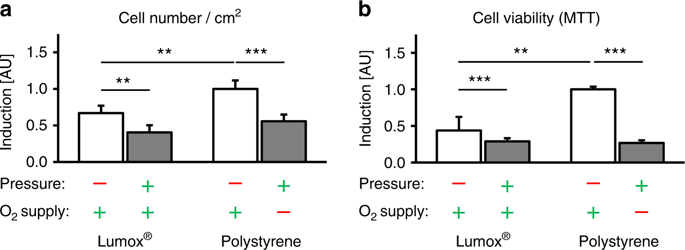
Determination of cell number and cell viability via MTT assays
The cell number per cm2 was determined after 72 h of incubation with a Beckman Coulter Counter Z2™ (Beckman Coulter GmbH, Krefeld, Germany). The cell viability of the hPDL fibroblasts was determined for all experimental groups by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays. For the final 5 h of the 72 h incubation phase, 400 µL of MTT solution in PBS (5 mg•mL–1, 4022.1, Carl Roth GmbH & Co. KG, Karlsruhe, Germany) was added per well. After the removal of the medium, 1 mL of DMSO per well was added. The hPDL fibroblasts were then incubated for another 5 min at 37 °C, and the absorbance was quantified at 550 nm by means of an ELISA reader (Multiscan GO Microplate Spectrophotometer, Thermo Fisher Scientific Inc., Schwerte, Germany), which corresponded to the cell viability.
Determination of the relative gene expression via quantitative real-time polymerase chain reaction
We quantified the expression of genes involved in inflammation (COX-2), collagen synthesis (COL1A2), angiogenesis (VEGF) and osteoblastogenesis (ALPL), and the expression of RANKL and its decoy receptor osteoprotegerin (OPG), which are the most important signalling molecules in the process of osteoclastogenesis.37
The RNA isolation and quality assessment was performed as described previously according to MIQE guidelines.44,45 Briefly, we isolated total RNA from hPDL fibroblasts by adding 1 mL of peqGOLD TriFastTM (PEQLAB Biotechnology GmbH, Erlangen, Germany) per well and performing the isolation according to the manufacturer’s instructions. The RNA pellet was eluted in 25 µL of nuclease-free water (T143, Bioscience-Grade, Carl Roth GmbH & Co. KG). The utilised extraction protocol ensured good RNA integrity (RIN, 28 S/18 S ratio) and the absence of genomic DNA and contamination, as shown previously.44 For the purity assessment and the quantification of the eluted total RNA, the optical density (OD) was photometrically measured at 280 , 260 and 230 nm (NanoDrop ND-2000, Thermo Fisher Scientific Inc.), with an OD260nm value of 1 representing 40 ng/µL total RNA. An OD260 nm/280 nm ratio of >1.8 indicated protein-free RNA, and an OD260 nm/230 nm ratio of >2.0 indicated phenol-/ethanol-free RNA.44
For cDNA synthesis, we used a standard amount of 1 μg of RNA per sample and transcribed it into cDNA (incubation for 60 min at 37 °C) by using 0.1 nmol of an oligo-dT18 primer (1 µL, SO131, Life Technologies, Thermo Fisher Scientific Inc.), 0.1 nmol of a random hexamer primer (1 µL, SO142, Life Technologies), 40 nmol dNTP mix (1 µL, 10 nmol per dNTP, Roti®-Mix PCR3, L785.2), 4 µL of 5× M-MLV-buffer (M1705, Promega, Fitchburg, WI, USA), 40 U (1 µL) of an RNase inhibitor (EO0381, Life Technologies) and 200 U (1 µL) reverse transcriptase (M1705, Promega) in 20 µL of nuclease-free H2O (Roth BioScience Grade T143, Carl Roth GmbH & Co. KG). After the heat inactivation of the reverse transcriptase (95 °C, 2 min), the first-strand cDNA was diluted 1:5 and stored until use at −20 °C. To minimise experimental variation, cDNA synthesis was performed for all samples at the same time.
For RT-qPCR amplification, we used the Mastercycler® ep Realplex-S thermocycler (Eppendorf AG, Hamburg, Germany). For each reaction, we mixed 7.5 µL of SYBR® Green JumpStart™ Taq ReadyMix™ (S4438, Sigma Aldrich®), 7.5 pmol (0.75 µL) of the respective primer pair (3.75 pmol per primer) and 1.5 µL of the respective diluted cDNA, and then added nuclease-free H2O (BioScience Grade T143, Carl Roth GmbH & Co. KG) to bring the total volume to 15 µL. To avoid technical errors during manual pipetting, all components except the cDNA solution were prepared as a master mix. cDNA amplification was performed with 45 cycles (initial heat activation 95 °C/5 min, per cycle 95 °C/10 s of denaturation, 60 °C/8 s of annealing and 72 °C/8 s of extension). The SYBR Green I fluorescence was quantified at 521 nm at the end of each extension step. The Cq values were determined as the second derivative maximum of the fluorescence signal curve with the software Realplex (version 2.2, Eppendorf AG, CalqPlex algorithm, Automatic Baseline, Drift Correction On). For the normalisation of the target genes (relative gene expression), we used a set of two reference genes (RPL22 and PPIB), which have been shown to be stably expressed in hPDL fibroblasts under the conditions investigated.44 The relative gene expression used for the statistical analysis was calculated as 2–ΔCq, with ∆Cq = Cq (target gene) – Cq (mean RPL22/PPIB), divided by the respective 2–ΔCq arithmetic mean of the Lumox® normoxic control group to set the relative gene expression to 1.15,16
We designed all primers (Table 1) according to the MIQE quality guidelines45 and previously described criteria44 by using NCBI PrimerBLAST and additional software to avoid the formation of dimers and secondary structures at the annealing temperature. The unmodified primers were synthesised and purified by Eurofins MWG Operon LLC (Huntsville, AL, USA; High Purity Salt Free Purification HPSF®). We performed a no-template control (NTC) without cDNA to assess possible faults resulting from primer dimers or contaminating DNA. The primer specificity was validated as described previously (melting-curve analysis and agarose gel electrophoresis).44
Enzyme-linked immunosorbent assays
For the quantification of OPG, soluble RANKL, ALPL, prostaglandin E2 (PGE2) and VEGF protein secretion in the hPDL cell supernatant, we used commercially available ELISA kits according to the manufacturers’ instructions (OPG: EHTNFRSF11B, Thermo Fisher Scientific Inc.; sRANKL: RD193004200R; Biovendor, Brno, Czech Republic; ALPL: OKEH00757; Aviva Systems, San Diego, USA; PGE2: 514010; Cayman Chemicals, Ann Arbor, USA; VEGF-A: RAB0507, Sigma Aldrich). We used cell culture supernatants from two independent experiments (N = 2) with a total of six biological replicates (n = 6). For the ELISA of OPG, we diluted the cell supernatants 1:10 in appropriate dilution buffer. The protein expression per well was related to the respective number of hPDL fibroblasts, as counted with a Beckman Coulter Counter Z2™ (Beckman Coulter GmbH).
Quantification of total collagen in the cell culture supernatant
For the quantification of total collagen, we used a commercially available kit (K218-100, Biovision, Milpitas, USA) according to the manufacturer’s instructions.
Quantification of RANKL and HIF-1α stabilization via western blot
Since RANKL can be expressed as two subtypes—soluble and membrane-bound—we also investigated the expression of membrane-bound RANKL by performing immunoblotting with a RANKL-specific antibody. In addition, we assessed the stability of HIF-1α, which, among other target genes, regulates COX-2 and VEGF expression.26,35 Total protein from hPDL fibroblasts was isolated with 100 µL of CelLytic™ M per well (C2978; Sigma-Aldrich®) supplemented with proteinase inhibitors (Carl Roth GmbH & Co. KG). To reduce proteinase activity, the proteins were kept on ice for the entire procedure. The determination of protein concentration was performed with RotiQuant (K015.3; Carl Roth GmbH & Co. KG) according to the manufacturer’s instructions. For immunoblotting, we separated equal amounts of total protein on a 10% SDS-polyacrylamide (RANKL) or 8% SDS-polyacrylamide (HIF-1α) gel under reducing conditions and transferred the proteins onto polyvinylidene difluoride (PVDF) membranes via electroblotting. To reduce the nonspecific binding of antibodies, we blocked the membranes with 5% nonfat milk in Tris-buffered saline and 0.1% Tween 20, pH 7.5 (TBS-T), at 4 °C overnight. Then, we incubated the membranes with anti-RANKL (1:2 000, ABIN500805, Antibodies-Online, Aachen, Germany), anti-HIF-1α (1:2 000, Santa Cruz Biotech, Heidelberg, Germany), anti-HSP90 (reference, 1:500, Santa Cruz Biotech) and anti-β-actin (reference, 1:5 000, Sigma-Aldrich®) for 1 h at room temperature. After washing three times in TBS-T, we incubated the blots for another 1 h with horseradish peroxidase-conjugated anti-rabbit IgG (Pierce, Rockford, USA) diluted 1:5 000 in 0.5% milk in TBS-T at room temperature. We visualised the antibody binding by using an enhanced chemiluminescence system (Pierce, Rockford, USA).
TRAP histochemistry (hPDL-mediated osteoclastogenesis)
To investigate the effect of mechanotransduction vs. that of hypoxia on the mediation of osteoclastogenesis by hPDL fibroblasts during orthodontic tooth movement, we performed coculture experiments with osteoclast-precursor cells. At the end of the total 72-h incubation period, hPDL fibroblasts from each experimental group were washed (PBS), and a macrophage osteoclast-precursor cell line (immortal murine RAW264.7 cells, CLS Cell Lines Service, Eppelheim, Germany) was added after force application at a concentration of 70 000 cells per well, thus avoiding the potential force-induced induction of RAW cell differentiation that was not mediated by RANKL.39 The resulting coculture was then incubated for another 72 h under specific cell culture conditions.6,39 Histochemical TRAP staining (red) was used to detect differentiated osteoclast-like cells.46 TRAP-positive cells were quantified at a magnification of ×100 with an Olympus IX50 microscope (Olympus, Germany) in ten random fields of view per well (biological replicates) by a blinded observer, and the arithmetic mean was used for further analysis.
Statistical analysis
Prior to the statistical analysis, all absolute data values were divided by the respective arithmetic mean of the Lumox® normoxic control group to obtain normalised data values relative to the values of the controls, which were set to 1. Using the software application SPSS® Statistics 24 (IBM®, Armonk, NY, USA), all data were tested for a normal distribution (Shapiro–Wilk test, visual assessment of histograms) and the homogeneity of variance (Levene’s test). The descriptive statistics are given as the mean (M) ± standard deviation (SD). The normal distribution of all data was confirmed. The experimental groups were compared by one-way ANOVA and validated by Welch’s test, since the homogeneity of variance was not always present. We used Games–Howell post hoc tests for heterogeneous variances in pairwise comparisons. Statistical significance was assumed at P ≤ 0.05.