Ethics statement
This study was approved by the Institutional Review Board (IRB) of the University Dental Hospital (Approval number: PNUDH-2019-009). The IRB of the University Dental Hospital waived the need for individual informed consent, and thus, a written/verbal informed consent was not obtained from any participant, as this study had a non-interventional retrospective design and all the data were analyzed anonymously.
Materials
In this study, images of 102 patients (aged 18–90 years) undergoing CBCT for TMJ diagnosis between 2008 and 2017 at the University Hospital were used. CBCT scans were performed using a PaX-Zenith 3D system (VATECH Co., Hwaseong, Korea) with 5.0–5.7 mA, 105 kV, a 24-s exposure time, a voxel size of 0.2–0.3 mm, and a field of view of 16 × 16 or 24 × 24. Patients presenting previous surgical history, malformation, or diseases of the oral and maxillofacial region were excluded.
Two trained researchers traced the mandibular canal in cross-sectional images using INVIVOTM (Anatomage, San Jose, CA, USA) dental imaging software to generate a ground truth image. For the practical annotation processing, the INVIVO’s cross-sectional view was annotated at 1 mm intervals, following the restoration of the original interval of 0.2 mm using 3D cubic interpolation. An oral and maxillofacial radiologist, with 6 years of experience, clarified the positions of any uncertain mandibular canals. The original image was stored with a tracing image. The tracing image was then replaced with the ground truth label that consisted of the mask for the mandibular canal (white) and background (black). The canal mask was extracted using the color information from the tracing image.
Methods
Preprocess
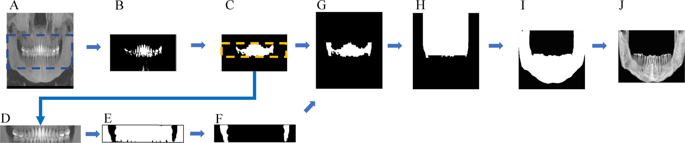
The size of all dimensions was resized by half before the preprocess. In order to increase accuracy and at the same time reduce the volume that the network learns, preprocessing was performed such that automatically only the 3D mandibular part from the raw data remained (Fig. 1). First, the center one-third of the reconstructed panoramic view (2D) (A) was binarized with teeth threshold (B) and then dilation was performed, leaving only the largest object (C). The resulting image, confined to the tooth height (D), was binarized with bone threshold (E). Leaving the two largest images after complementing the image (F), a buccal corridor between the ramus and jaw bone was obtained. Next, this buccal corridor and the tooth part (C) was combined (G), the maxillary part was obtained by extending the image (H) upwards. The maxillary region was removed from the 3D binarized jaw bone image, and a 3D closing operation was performed to obtain a binarized mandibular image (I). After confining the area within the bounding box of this image, the mandible image was finally obtained, thus leaving only the inner part of the mask in the original image (J). In (A) and (D), the binarization coefficients of the teeth and bone were accurately calculated by limiting the area, and the binarization coefficients were obtained using the multi-level Otsu’s method. The bone threshold used the first level and the teeth used the third level threshold. In the case of inaccurate mandibular segmentation result, the threshold of the bone and tooth were manually readjusted.
Preprocessing steps such that just the mandibular part remained.
Networks
2D Networks
First, two image segmentation architectures, U-Net and SegNet, that share similar encoder and decoder network architectures, except for some differences, were implemented. SegNet uses the basic architecture from VGGNet20 with the pre-trained convolutional layer and batch normalization, while its decoder uses the max pooling indices to up-sample the feature map instead of learning like Fully Convolutional Network (FCN)21. With dental images, the same number of filters were used as illustrated in Fig. 2A. In contrast, U-Net had up-sampling operators by learning to deconvolute the input feature map and combine it with the corresponding encoder feature map for a high-resolution feature map as the decoder output.
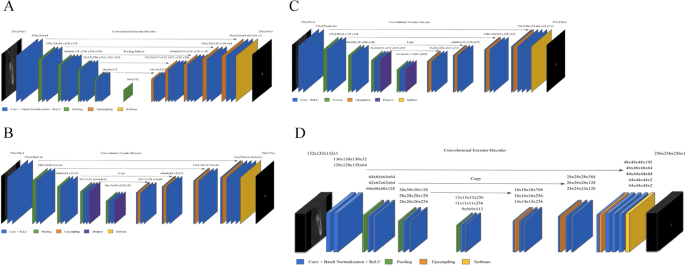
Architecture of deep learning networks. (A) SegNet; (B) U-Net with fewer filters than the original U-Net (C) U-Net with the original number of filters; (D) 3D U-Net.
In this study, the original architecture of 2D U-Net was modified as follows: first, the feature maps were padded with zeros using the ‘same padding’ instead of the ‘valid padding’ in the convolutional layers, so that an input image was fully covered by specified filter and stride; next, the cropping process was removed before copying the drop-out or convolutional layer outcomes as shown in Fig. 2B,C. Even though the valid sampling and cropping processes were key points originally used in the U-Net algorithm to find the hidden pattern and convey to the deep network, in this study, the number of pixels of the desired detection area (mandibular canal) was very small and was located partially on the edge. Hence, padding feature maps filled with zeros were applied to maintain the same dimensions, to avoid over-fitting via class imbalance and information loss of the edge surrounding the mandibular canal at the corner22. The 2D U-Net was first studied with both small and original number of filters as the original U-Net and SegNet. The 2D U-Net architecture with original filters was then experimented with pre-trained weights from 2D VGG net. Additionally, 2D U-Net pre-trained from 2D VGG net was examined with 4 adjacent images. Given the fact that doctors see adjacent images when diagnosing 3D images, this network structure was expected to obtain more contextual information from 2D adjacent images and simultaneously circumvent the computation burden of 3D training.
3D network
A 3D U-Net18 fully convolutional network for 3D canal segmentation was also used. It is an extension of the 2D U-Net layers in 3D (Fig. 2D), which was learned by randomly selecting 64 3D patches with each image of the size 132 × 132 × 132 pixels. The 3D U-Net used the same 2D U-Net architecture, with corresponding 3D operations (3D convolutions, 3D max pooling, and 3D up-convolutional layers)18, batch normalization addition, and dropout layer removal. Since 3D network gets more contextual information, it maintains valid padding as its original 3D architecture.
Training options
With binary cross entropy, class weight of 5.3:1000 was used in all networks to compensate class imbalance by using the pixel label counts. We used median frequency balancing23 for calculating the class weight as proposed in SegNet. Of the 49094 images, the dataset was divided into train:valid:test sets with the ratio of 6:2:2, and train:valid:test datasets had equal class images. Each image originally had 545 × 900 pixels, and it was used as 256 × 256 pixels for 2D and 132 × 132 × 132 pixels for 3D. NVIDIA Titan RTX GPU with cuDNN version 5.1 acceleration was used for 3D network training.
2D network
The U-Net and SegNet were trained with and without a pre-training class weight individually. The U-Net was first studied with 1) fewer filters than retaining the original U-Net architecture and 2) larger number of filters (deeper network) of the original U-Net and SegNet. Moreover, with the original SegNet architecture, pre-trained weights from VGG net could be used not only for SegNet but also for U-Net. Therefore, U-Net and SegNet with 1 image, U-Net with 4 adjacent images were studied with transfer learning. The models were trained for 600 epochs with Adam optimizer24 with a learning rate and decay (0.01, 0.005) for SegNet and (0.0001, 5e−4) and a momentum of 0.9 for U-Net instead of 0.99 from U-Net’s original proposed momentum. We trained the 2D variants until the training loss converged. The model with the best performance on a validation dataset was selected.
3D network
The models were trained for 100 epochs using Adam optimizer24 with a learning rate of 5e−4 decayed by a factor of 5 after 5 epochs, and a batch size of 8.
Metrics for accuracy comparison
The canal and background pixel accuracy, global accuracy, class accuracy, and mean IoU (intersection over union) were assessed to evaluate the accuracy. Each definition is as follows:
$${rm{Pixel}},{rm{accuracy}},{rm{of}},{rm{canal}}=frac{{rm{TP}}}{{rm{TP}}+{rm{FP}}}$$
$${rm{Global}},{rm{accuracy}}=frac{{rm{TP}}+{rm{TN}}}{{rm{TP}}+{rm{TN}}+{rm{FP}}+{rm{FN}}}$$
$${rm{Class}},{rm{accuracy}}={rm{average}},{rm{of}},{rm{pixel}},{rm{accuracy}},{rm{of}},{rm{canal}},{rm{and}},{rm{background}}$$
$${rm{IoU}},{rm{of}},{rm{canal}}=frac{{rm{TP}}}{{rm{FN}}+{rm{TP}}+{rm{TN}}}$$
$${rm{Mean}},{rm{IoU}}={rm{average}},{rm{of}},{rm{IoU}},{rm{of}},{rm{canal}},{rm{and}},{rm{background}}$$
$${rm{TP}}:{rm{true}},{rm{positive}},{rm{FP}}:{rm{false}},{rm{positive}},{rm{FN}}:{rm{false}},{rm{negative}},{rm{TN}}:{rm{true}},{rm{negative}}$$